www.fluorescencemicroscopy.it
Main menu:
- Home Page
- Microscopes
- Fluorescence
- Description
- Diascopy
- Epi-Fluoresc.
- Illumination
- Installing and Aligning a Mercury Lamp
- Fluorocromes
- Tables of Fluorocromes
- The Fading
- Wavelengths
- Properties
- % of transmission
- Types of filters
- Overlapping Spectra
- Intensity
- Image acquisition
- The Filter's set
- The Filters
- Fluorochrome's praparation
- Sample's preparations
- Fluorescence Sample
- Cytochemical Fluorescence
- Intrinsic Fluorescence
- Gallery
- Phase Contrast
- Polarization
- Darkfield
- DIC
- COL
- Rheinberg
- Brightfield
Fluorescence Sample
Fluorescence
FLUORESCENCE SAMPLE
Cheek epithelial cells
Use a wooden toothpick and gently scrape the inside of your cheek. Apply the extracted epidermal cells to a small drop of water plus fluorescent dye.
Throat swab
Use a QTip to take a palate swipe. Apply the sample to a small drop of water/dye solution on a microscope slide. Apply a coverslip and secure with fingernail polish.
Elodea leaf
This common aquarium plant is an excellent sample for light microscopy. The cells containan array of cytoplasmic strands, nuclei, and visible organelles. In addition, there is a rich population of epiphytes growing on the leaf surface. Gently place a leaf into a small drop of water, probe, or fluorescent dye on a microscope slide. For cell wall staining a 5 sec dip in Hexane helps dye penetration.
Plants
Use a razor blade to cut small sections of plant material. The sections should be <3mm on a side, and as thin as possible. A common sample is the inner epidermis of onion. Cut a deep square in an onion, pull out three layers (storage leaves), cut a small square on the surface of the inner leaf. Peel it off with a forceps and place immediately in a drop of water with probing dye.
Microbial specimens
Simply place the collected sample in a small drop of water, probe, or fluorescent dye. Possible collection methods:
- Soil sample
- Surface sample (using toothpicks or tape)
- Biofilm samples from sink drains
- Throat/Tongue swabs
- Tooth tarter samples
- Yogurt or cheese. Good source of microorganisms Lactobacillus acidophilus, L. bulgaricus,
- and Streptococcus thermophilus.
- ProGreens (mixture of bacteria and cyanobacteria). Mix with water, incubate >1h,
- centrifuge. Use the supernatant.
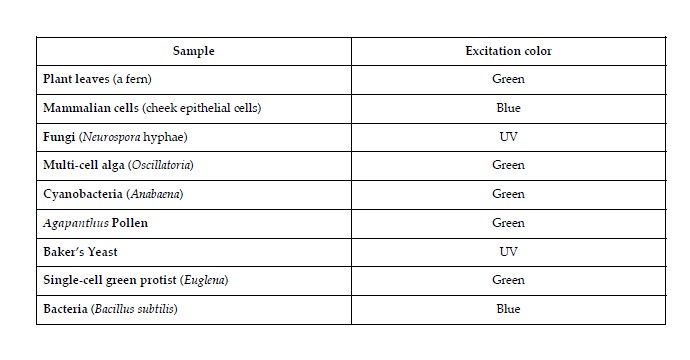
TESTING SAMPLE
TESTING LIQUIDS FOR BACTERIAL CONTAMINATION
For testing liquids for bacterial contamination must be used a disposable filtration unit. The unit fits onto a syringe. Aqueous solutions, with putative bacteria is pushed through a small (13mm) 0.2ìm glass filter, which catches the bacteria, supported by a larger paper filter. The glass “Anodisc” filter is recovered and mounted on a glass microscope slide. It is small enough to fit under a standard 22mm square microscope coverglass. Bacteria will be stained with the fluorescent nucleic acid dye SYTO BC to make them visible on the filter surface.
IDENTIFYING BACTERIA ON SURFACES
Identifying bacteria on surfaces is a procedure that can be performed quickly and easily using fluorescent probes previously discussed. The general procedure is to determine the intrinsic fluorescence of the surface in question, then choose a dye with a fluorescence spectrum that differs from background. Probe the sample with SYTO BC to localize the bacteria by fluorescence microscopy.
DETERMINING BACTERIA VIABILITY USING FLUORESCENCE MICROSCOPY
Bacterial cell viability can be determined using fluorescence microscopy, and the fact that in living cells certain dyes are excluded by an intact outer cell membrane. Can be used two DNA dyes: SYTO and Propidium Iodide (PI) to determine the state of the plasma membrane, and thus whether the cells are living or dead. Both dyes enter a dead cell (having a disrupted or absent membrane), whereas PI is excluded by the membrane of a living cell. SYTO freely crosses the membrane and stains DNA in living cells. When used in combination, living cells fluoresce green because they are stained only with SYTO, whereas dead cells fluoresce red when stained by both dyes. The red fluorescence of PI quenches green fluorescence of SYTO.
BACTERIAL ENDOSPORE STAINING
Certain bacteria genera (Bacillus, Clostridium) develop resistant endospores during periods of environmental stress. These spores are highly resistant to desiccation and UV damage, and because of this they also are resistant to the normal methods of fluorescence staining. Can be used a method adapted from cytology experiments in plants to modify the endospore cell wall sufficiently to introduce the fluorescent dye Acridine Orange.
FLUORESCENT INDICATOR OF VIABILITY—CALCEIN AM OR FLUORESCEIN DIACETATE
Calcein AM and Fluorescein diacetate (FDA) are membrane-permeant non-fluorescent dyes that can be
introduced into cells via incubation. Once inside the cells they are hydrolyzed by endogenous esterase (plus ATP) into the negatively charged fluorescent calcein or fluorescein. Thus, these dyes can be used as indicators of metabolism, and therefore cell viability.
FLUORESCENT DYE FOR CHROMATIN— PROPIDIUM IODIDE (PI)
Propidium Iodide is a membrane-impermeant nucleic acid intercalator. The dye is commonly used to selectively stain dead cells in a cell population and also used as a counterstain for nuclei in multicolor fluorescent imaging.
FLUORESCENT INDICATOR OF BACTERIA— PHLOXINE B
Phloxine B, a red acid dye, is a derivative of fluorescein with distinctly bluish shade; used for disinfection and detoxification of wastewater through photo-oxidation (it binds to bacteria). It is used as an intermediate for making photosensitive dyes and drugs. It is used as a cytoplasm stain in histology. Phloxine stains bacteria in mixed samples. Phloxine photobleaches easily.
PLANT AND FUNGI CELL WALLS
The cell wall of these organisms consists of the polymer of glucans. The glucan found in plant cell walls is cellulose. The glucan found in fungi is chitin. Cellulose and chitin are not natively fluorescent, however the fluorescent probes Calcofluor and Acridine Orange may be used to locate and identify these components, and indirectly cell walls.
LIPIDS
All living organisms contain lipids (usually sequestered within organelles). Lipids generally do not fluoresce however, so these components are candidates for simple fluorescence probing using the fluorescent dye Nile Red. This dye is lipophilic and thus will be localized to lipid-containing organelles (liposome) in cells. When excited by blue light, Nile Red fluoresces yellow/gold.
IDENTIFYING BACTERIA IN EUKARYOTIC CELL PREPARATIONS
Eukaryotic cells (animal and plant) can be used to gain a familiarity with the morphology, staining ability, and size of bacterial cells. These cells have DNA-containing nuclei, so DAPI and/or PI are good fluorescent probes to use. These dyes may also be used to identify the bacteria cells in the surrounding neighborhood.
IDENTIFYING BACTERIA USING A SPECIALIZED FLUORESCENT PROBE
One of the initial tests in an investigation of an unknown sample is to determine the presence of bacteria.
Fluorescence probing for DNA is one positive method of identifying the presence of bacteria. Molecular Probes’ Bacteria Counting (SYTO BC; B-7277) stain is a high-affinity nucleic acid stain that easily penetrates both grampositive and gram-negative bacteria and results in an exceptionally bright green fluorescent signal. The Molecular Probes kit is designed for flow cytometric analysis of bacterial populations, however, the SYTO BC dye is useful for microscopy investigations.
The criteria used for microscopic identification of prokaryotes include cell shape, size, grouping, Gram-stain reaction, and motility. Bacterial cells almost invariably take one of three forms: rod (bacillus), sphere (coccus), or spiral (spirilla and spirochetes). Bacilli may occur singly or form chains of cells; cocci may form chains (streptococci) or grape-like clusters (staphylococci); spiral shape cells are almost always motile; cocci are almost never motile.
The following table shows the results obtained by using different sets of filters for each preparation. For each test carried out shows the type of set (dualband or single-band), the bandwidth of excitation and emission filter, the result obtained on the various components of the cell and not, taken into consideration. They were considered some of the main constituent parts of the cell, and in any event that will deliver greater assurance of marking: nucleus, cytoplasm, mitochondria, lysosomes, centrosomes, microtubules, actin. They are part of the test, as external components, bacteria. The color shown in the description of the component is obtained from the same component in the test phase of the "table"results. The assigned value, indicates as was appropriate to use this filter set up with that type of sample and using that particular fluorophore. The highest value indicates the best result.
FLUOROCHROME |
EXC |
EMIT |
RESULTS OF TESTS ON HUMAN CHEEK CELLS BASED ON THE FILTER SET USED |
HOECHST 3258 |
346 |
460 |
B/R = Nucleus, Lisosome, Cytoplasm, Microtubules, Bacteria, R=5 |
Acridine Orange (+RNA) |
460 |
650 |
G/R = Nucleus , Lysosome, Cytoplasm, Microtubules, Bacteria, R=5 |
BO (Thiazole Yellow) |
460 |
480 |
G/R = Nucleus, Lisosomes, Cytoplasm, Bacteria, R=5 |
BOBO-1 |
462 |
481 |
G/R = Nucleus, Lisosome, Cytoplasm, Centrosomes, Microtubules, Bacteria, R=5
|
Dye 307 |
485 |
590 |
G/R = Nucleus, Lisosome, Cytoplasm, Centrosomes, Microtubules, Bacteria, R=4
|
Nile Red |
485 |
525 |
G/R => Nucleus,Vacuoesi, Cytoplasm, Mitochondria, R=3 |
Fluorescein |
494 |
518 |
FITC = Nucleus, Cytoplasm, Vacuoles, Actin, R=3 |
Carboxyfluorescein |
492 |
518 |
|
Acridyne Orange (+DNA) |
500 |
526 |
G/R = Nucleus, Lisosome, Cytoplasm, Microtubules, Bacteria, R=5
|
Rhodamine G |
502 |
527 |
|
TOTO-1 |
509 |
533 |
G/R = Nucleus, Lisosome, Cytoplasm, Centrosomes, Microtubules, Bacteria, R=5
|
Thiazole Orange |
509 |
530 |
G/R = Nucleus, Lisosome , Cytoplasm, Centrosome, Microtubules, Bacteria, R=4
|
Heptyl-thiazole Orange |
515 |
535 |
|
N-desmethyl- thiazole Orange |
515 |
535 |
|
Carboxyeosin |
515 |
542 |
|
EOSIN |
524 |
544 |
G/R => Nucleus, Mitochondria, Cytoplasm, Microtubulus, R=3
|
Erythrosin |
529 |
544 |
G/R => Nucleo,Vacuoli, Citoplasma, R=2
|
Rhodamine B |
555 |
580 |
|
Carboxyrhodamine B |
556 |
581 |
|
P2 (Pyrillium) |
580 |
640 |
G/R = Nucleus, Lisosomi, Cytoplasm, Centrosomi, Microtubules, Batteri , R=4
|
G/R Dualband ..................B/R Dualband...............DAPI...............................FITC...............................TRITC
EXC 488 EMI 528...............EXC 389 EMI 460...........EXC 330 EMI 450................EXC 485 EMI 530................EXC 535 EMI 605
EXC 580 EMI 630...............EXC 530 EMI 570
Sub-Menu:
- Description
- Diascopy
- Epi-Fluoresc.
- Illumination
- Installing and Aligning a Mercury Lamp
- Fluorocromes
- Tables of Fluorocromes
- The Fading
- Wavelengths
- Properties
- % of transmission
- Types of filters
- Overlapping Spectra
- Intensity
- Image acquisition
- The Filter's set
- The Filters
- Fluorochrome's praparation
- Sample's preparations
- Fluorescence Sample ←
- Cytochemical Fluorescence
- Intrinsic Fluorescence
- Gallery